|
|
|
| Tools |
>> Alignment Analysis |
 |
Multiple Sequence Alignment Tool.
|
 |
| |
ESPript: Easy Sequencing in Postscript |
 |
Generate a pretty PostScript output from aligned sequences. Obtain coloring and residue grouping in the output from a similarity score calculated following several optinals algorithms. Take a PDB file with the 3D-coordinates of the first sequence in the aligment, in which case the ouput is enhanced with the secondary structure, solvent accessibility, and hydropathy.
|
| |
ENDscript: Analyse and generate multiple output from a PDB |
 |
An interface grouping seven programs: BLAST; MULTALIN/ CLUSTALW; DSSP; CNS; ESPript; BOBSCRIPT/ MOLSCRIPT; PHYLODENDRON. Aims to produce from a single PDB file, three PostScript figures containing most required sequence and structure information on a protein. Just upload your PDB and hit the RUN button.
|
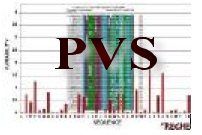 |
A server which calculates the variability from multiple sequence alignment using the Shannon entropy (H) function, the Simpson entropy function and the Wu-Kabat entropy function.
|
 |
SeqLogo generate sequences logos from amino acid sequence alignment. Sequences logos are useful tools to visualyze sequence patterns and represent a more informative alternative to consesus sequence.
|
 |
This tool calculates pairwise identity and similarity of a msa using several methods.
|
|
 |
|
 |
|
|
| |
|
|
|
|