ESPript, Easy Sequencing in Postscript, is a utility to generate
a pretty PostScript output from aligned sequences.
Reference article is: Gouet, P., Courcelle, E., Stuart, D.I.
and Metoz, F. (1999). ESPript: multiple sequence alignments in PostScript.
Bioinformatics.
15 305-8
Key Features
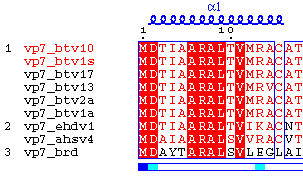
ESPript is a utility, whose output is a PostScript file of aligned
sequences with graphical enhancements. Its main input is an ascii file
of pre-aligned sequences. Optional files allow further rendering. The program
calculates a similarity score for each residue of the aligned sequences.
The output shows:
-
Secondary Structures
-
Aligned sequences
-
Similarities
-
Consensus
-
Accessibility
-
Hydropathy
-
User-supplied markers
-
Intermolecular contacts
In addition, similarity score can be written in the bfactor column of a
pdb file, to enable direct display of highly conserved areas.
Installing standalone ESPript
1.Fill the licence form
- License form for academic users to be
faxed or to be sent by paper mail to the authors
- Licence form for commercial user
to be sent by registered letter with acknowledgement of receipt to the CNRS
(see article 13 of the form). A sum of 1000 euros is requested within 35 days
(see article 7 of the form).
3.THEN dowload the program
The source ESPript.f is a one-file FORTRAN program, available via
our download page.
You can compile the source by using the Makefile. Just type
:
make
By default, the binary is placed in the bin directory. The
program has been tested under Unix and Linux.
ESPript can be executed using a standard input.
The distributed version (check releases) is configured for a maximum of 300 sequences with residues
aligned on 2,000 columns. This default can be changed in the fortran source.
Scripts for the html interface described below are included in the distributed tar file.
Running ESPript via the web
You can run ESPript from this server with the html interface.
It is configured for a maximum of 1,000 sequences. Click on the red button
below and fill the form. Then, you just have to click to receive, and possibly
display, the output PostScript file and optional pdb file. You can also
convert the PostScript to GIF or TIFF files via the interface (this uses
an external conversion program, for instance the program convert).
You may have to configure your browser with appropriate helpers applications
to display the generated output. The mime type of the postscript file is
application/postscript,
it could be linked to ghostview.
The mime type of the pdb file is chemical/x-pdb, it could be linked
to rasmol.
If you want to install the script of the html interface on your
server (Unix or Linux-Apache), please read the installation
documentation.
The cgi-bin interface is covered by the gnu public license.
Main site is
Laboratoire de BioCristallographie
, Institut de Biologie et Chimie des Protéines (CNRS UMR 5086) - Lyon, France.
Mirror sites are Columbia
University Bioinformatics Centre, New York,
USA and
Bioinformatics Centre,
Singapore.
Links to webESPript
ENDscript:
you can upload a PDB file or enter a PDB code such as 1M85. The programs DSSP and CNS are executed via the interface, so
as to obtain an ESPript figure with a lot of structural information
(secondary structure elements, intermolecular contacts). You can
also find homologous sequences with a BLAST search, perform multiple sequence alignments with MULTALIN or CLUSTALW and create
an image with BOBSCRIPT or MOLSCRIPT to show similarities on your 3D structure.
ProDom:
you can enter a sequence identifier to find homologous domains, perform
multiple sequence alignments with MULTALIN and click on the link to ESPript.
Predict
Protein: you can receive
a mail
in text (do not use the HTML option when you submit your request in Predict Protein) with aligned sequences and numerous
information including secondary structure prediction.
Click on a special html
link to upload your mail in ESPript.
NPS@: you can execute the programs BLAST and CLUSTALW
to obtain multiple alignments. You can predict secondary structure elements and click
on the link to ESPript.
ESPript: the calendar
Acknowledgements
This program started in the laboratory of Dr Richard
Wade at the Institut de Biologie Structurale,
Grenoble. It moved later to the Laboratory
of Molecular Biophysics in Oxford, then to the
Institut
de Pharmacologie et de Biologie Structurale in Toulouse.
It is now developed in the Laboratoire
de BioCristallographie of Dr Richard Haser, Institut de Biologie
et de Chimie des Protéines,
Lyon
and in the
Laboratoire
de Biologie Moléculaire et de Relations Plantes-Organismes,
group of Dr Daniel Kahn, Institut
National de la Recherche Agronomique de Toulouse.
Authors
ESPript was created
by Patrice Gouet and Frédéric
Metoz.
Emmanuel Courcelle
wrote the html-user interface.
Xavier Robert performed installation and administration of the server in Lyon.
The calendar is a masterpiece of Michel Gouet.
Send your messages to espript@ibcp.fr,
if you have questions, suggestions or if you want to be informed about
program releases.