USER GUIDE
E-mail
E-mail is optional. If you enter a valid e-mail address you will be notified once the job is ready,
otherwise you will be redirected to the result waiting page that will refresh until your results are ready.
Input
Input query for TAPREG can either be a protein sequence or one or several epitope sequences (epitopes length between 8 and 16) both in FASTA format, depending on
the option selected.
- Protein
Example of a protein sequences in FASTA format:
>gi|82503281|ref|YP_401725.1| BDLF3.5 [Human herpesvirus 4 type 1]
MSAPGCSERQDKKRGTIGEREFGELLSWDPTDLPRTVARVYVAVGGLFEQEVSEVQRLENICTLLDLAGVECQTKAD
- Epitope(s)
Example of several peptide sequences in FASTA format:
>peptide1
FMPKVNFEV
>peptide2
VVFLHVTYV
>peptide3
LPRPDTRHL
>peptide4
VFSDMETKVDTR
>peptide5
PYKRIEELLMPVGGQ
- Threshold
If a threshold is selected, only score values lower than the threshold will be shown.
- Models
Models were based on support vector machine regression algorithms that were trained on a set of 613 9-mer peptides of known affinity to TAP (logIC50relative).
Two models are available:
- DS_613 Sparse
Models was generated from sparse representations of input peptide sequences.
- DS_613 Blosum
Models was generated from blosum representations of input peptide sequences.
- Upload
Input query can be pasted or uploaded from a local file. If a local file is uploaded,
it will be processed by default. Files uploaded onto the server must contain only ascii characters.
In other words, make sure you save your files as TEXT before you make any attempt of uploading them onto this server.
Ouput
- Protein
The program splits the input protein sequence in all posible 9-mers, and computes tap score of
each of these partial sequences. The lower score, the highest the binding affinity.
Results are shown in a table sorted by tap score. The partial sequence, the initial position of
the sequence inside the protein and the tap score are provided, as shown below.
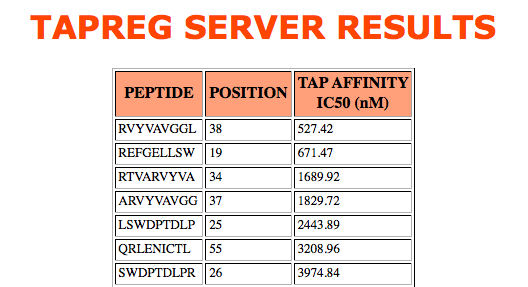
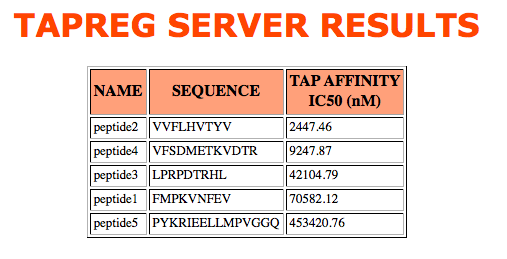
- Peptide(s)
Both, score and sequence provided, are shown in the result table, sorted by tap score.

Last change: March 2009

